NYU MRI Biophysics Group
Bridging the micro-macro gap with diffusion MRI
WHAT WE DO
Advancing MRI Imaging
We develop novel imaging markers based on advanced yet clinically usable MRI methods and biophysical modeling, as well as their validation and traslation into clinical applications. We are part of the Center of Biomedical Imaging, Department of Radiology at the New York University Grossman School of Medicine.
WHO WE ARE
A multidisciplinary team

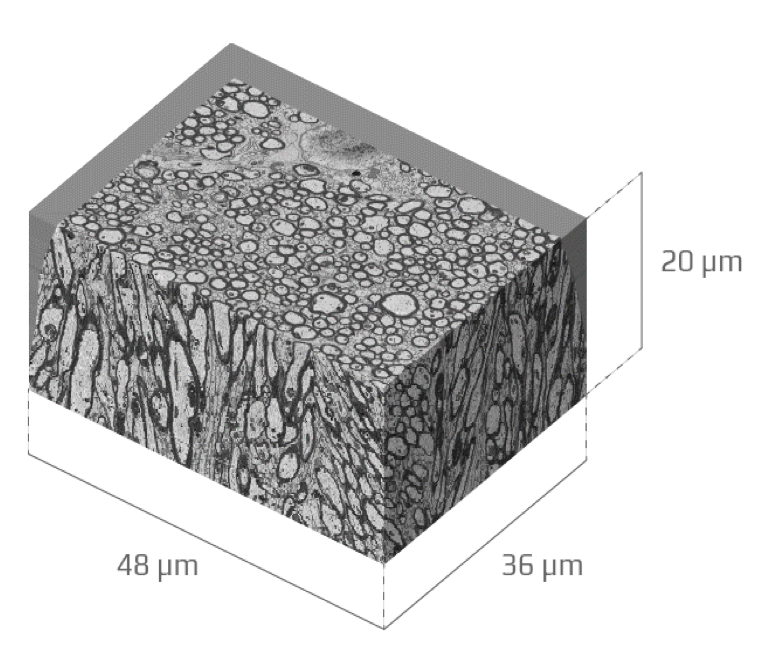
OUR WORK
Latest Publications
Resources
Our group is committed on contributing developed software to the scientific community by making it freely available on Github, and the open source MRTRIX.
SOFTWARE
ARTIFACT CORRECTION
The correction of the different types of artifacts (Gibbs ringing, EPI distortion, eddy current distortion, motion, etc) as well as noise is necessary for an accurate, precise and robust estimation of diffusion model parameters.
- DESIGNER (Diffusion parameter EStImation with Gibbs and NoisE Removal), a postprocessing pipeline for diffusion MRI images
- Denoising using Marchenko-Pastur Principal Component Analysis (MP-PCA), available on mrtrix.org (function _dwidenoise_) or as matlab code
- Outliers detection, available on visielab.uantwerpen.be/irlls
DIFFUSION PARAMETER ESTIMATION
- DTI/DKI processing, available on github.com/NYU-DiffusionMRI/Diffusion-Kurtosis-Imaging (functions DKI_fit.m, DKI_parameters.m)
- White Matter Tract Integrity metrics derived from DKI, available on github.com/NYU-DiffusionMRI/Diffusion-Kurtosis-Imaging (function wmti_parameters)
- Random Permeable Barrier Model (RPBM): MATLAB code for modeling restriction size and permeability, is available on github.com/NYU-DiffusionMRI/RBPM
AFFILIATIONS
Our Funding Partners




JOIN US
We are now recruiting
We are always looking for talented and passionate scientists who are interested in moving our research forward.
View our detailed job description here.